A just released paper in PNAS that reconstructs the mitochondrial DNA of a >300,000 year old cave bear lineage is getting some attention…and for good reason (Dabney, et al., 2013). You might wonder who cares about ancient cave bear lineages outside of ancient cave bear experts. But when those cave bear remains come from the site of Sima De Los Huesos, part of the Atapuerca archaeological complex, and home to the densest concentration of Middle Pleistocene hominin fossil remains anywhere in the world….well, then it gets more interesting.
To step back a bit…we know that DNA tends to preserve better in colder environments, particularly permafrost environments, where the rate of DNA degradation is diminished. Slower degradation means larger fragments of DNA persist for longer periods of time, making it easier to extract and read that DNA. The successful reconstruction of mtDNA from this more than 300,000 year old cave bear specimen at Atapuerca potentially triples the available time frame for looking at ancient human DNA, and was the product not of improved extraction or amplification methods, but instead of better utilization of genomic reference library techniques. Amplifying very small fragments of DNA using PCR requires primers to identify specific fragments, which limits the length at which you can effectively read. The genomic library methods outlined by Dabney, et al. try to circumvent this problem.
the possibility remains that not all sequence information residing in ancient specimens is optimally recovered with these methods. This possibility becomes apparent when inspecting the size distributions of sequences reported from ancient DNA (e.g., refs. 3, 8, and 15), which consistently show a mode of ∼40 bp or larger. It is unclear whether the deficit of shorter molecules is due to poor preservation in ancient biological specimens or their exclusion during sample preparation. This question is of importance because it is expected that the number of DNA fragments in an ancient sample increases exponentially as length decreases and, hence, that most information resides in very short molecules (3, 8). (emphasis added)
If we can properly identify the information available in short sequence reads, we are likely to get access to vastly more ancient genetic information than is currently available.
And obviously if the authors can achieve this with the Sima cave bears, maybe they can do it with the Sima hominins…
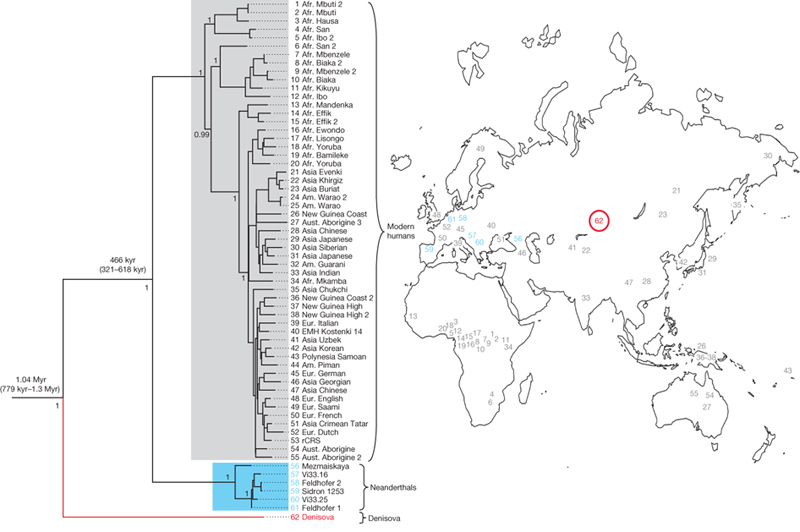
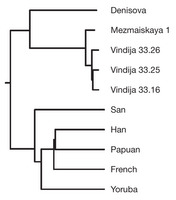
Which raises the question of why this is important and what we might expect to find. We already know that mtDNA is only a small part of the evolutionary and biogeographic story of ancient populations, with nuclear DNA presenting a more significant technical challenge to read (because you have vastly more copies of mtDNA than nuclear DNA in your cells, and mtDNA is structurally simpler and much smaller), but also much more valuable information. Current understandings of ancient mtDNA place Neandertals as a side branch relative to contemporary humans, with the mtDNA from Denisova Cave in Sibera a further outgroup (Reich, et al., 2010). Where mtDNA from Sima would fit in (if it is available), particularly relative to the relationship between Neandertals and Denisovans would be a fascinating, if not complete look at the phylogeography of these lineages.
John Hawks picks up on a further interesting aspect of the cave bear mtDNA itself:
Personally, I can’t wait until we have a thicker sampling of the Middle Paleolithic time slice for a number of species, because that will enable us to understand the population dynamics in response to at least two and possibly more glacial cycles in Europe.
The more comparative models we have for ancient DNA in other species, the better understanding we can have about how to interpret aDNA in humans from the climatically volatile Late Pleistocene. Indications of widespread regional extinction in other large-bodied mammals, for example, might provide good reason to refine the expectations and thereby test hypotheses about such phenomena in human prehistory. I will add, such data might also allow us to potentially begin to get around issues of equifinality by providing independent lines of evidence. For example, maybe aDNA evidence might support parallel reconstructions of population history in Neandertals and other large-bodied European mammals during the Riss glaciation period (~140-200 kbp), but contrasting models in the Würm period (~20-90 kbp). A differential response between humans and other species during this latter glacial period could allow a more direct test of whether or not archaeological changes in the latter Neandertal record are a result of environmental forcing or biocultural innovation and adaptation.
The greatest obstacle for hypothesis testing in the paleo-record is equifinality. Lots of possible answers seem equally likely (or at least indistinguishably likely) for a given question. Was it environment? Was it culture? Was it demography? Single lines of evidence–archaeological samples, fossil remains, aDNA–are insufficient to get around this problem. But coupling different lines of evidence, particularly when the predictions for such lines of evidence diverge, is, in my view, the most valuable aspect of the continuing emergence of ancient DNA Studies.
*****
1. Dabney, et al. (2013) “Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments” PNAS www.pnas.org/cgi/doi/10.1073/pnas.1314445110
2. Krause, et al. (2010) “The complete mitochondrial DNA genome of an unknown hominin from southern Siberia” Nature 464, 894-897. doi:10.1038/nature08976
3. Reich, et al. (2010) “Genetic history of an archaic hominin group from Denisova Cave in Siberia” Nature 468, 1053–1060. doi:10.1038/nature09710