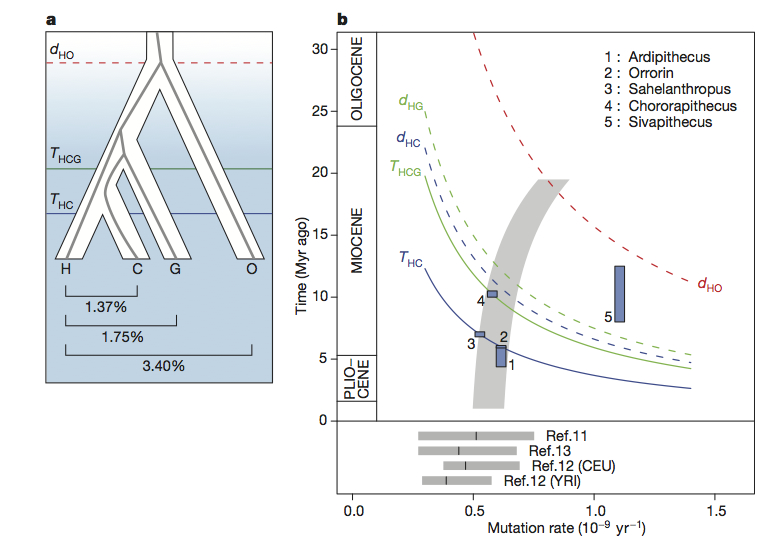
Nature published the results from the first full-genome sequencing of Gorilla this week. As with any new genome sequencing project, this initial publication is the result of an immense amount of work (the author list on the paper runs to 71 if I counted correctly), but is really only the starting point for asking interesting questions. After going through some of the technical specifications of the sequencing work, the first half of the Nature paper deals with the issue of estimating divergence times between humans, Pan and Gorilla given estimated demographic scenarios and mutation rates. This is an area with much significance for interpretations of the fossil record, but the authors decision to basically fit the data to a given interpretation of the fossil record means that the current interpretation does not add a lot of new value to questions as to where Ardipithecus, Orrorin or Sahelanthropus fit within the hominid lineage, for example (see figure b below).
The Nature News piece that accompanies the paper’s publication draws attention to evidence for positive selection on genes associated with hearing, an issue also discussed by John Hawks.
Personally, what I find most interesting is what the genome reveals about our understanding of speciation processes in large-bodied hominids. With the Gorilla genome joining the genomes of Homo sapiens, Pan troglodytes (chimpanzees), Pongo pygmaeus (Orangs) as well as the Neandertal and Denisovan genomes, we now have a dense assemblage of closely related extant and extinct taxa. And, not surprisingly, speciation in this group is a messy and complex process. This paper estimates that 30% of the Gorilla genome reflects incomplete lineage sorting (ILS), meaning nearly a third of the genome reveals closer evolutionary affinities between gorilla and chimps or gorillas and humans. We know that gorillas are actually an evolutionary outgroup to ourselves, chimpanzees and bonobos, so what the high degree of ILS reflects is the “fuzziness” associated with the process of speciation. Speciation in large-bodied mammals is not instantaneous and true isolating mechanisms are especially slow to develop, allowing for continued periodic or isolated instances of meaningful genetic exchange between diverging lineages. The authors provide a second good example of this in the text by suggesting the divergence between Eastern and Western gorilla species began close to 2 million years ago (a date, by the way, that seems to come up a lot), but with more recent genetic exchange between the lineages.
We also compare the western and eastern gorilla species, estimating an average sequence divergence time 1.75 million years ago, but with evidence for more recent genetic exchange and a population bottleneck in the eastern species.
The fundamental messiness of speciation across this group as evidenced by these genomic data is a big part of why I am not sympathetic to models of human evolution postulating many concurrent hominin taxa.
UPDATE: The study’s lead author, Aylwyn Scally, raises some clarifying points in the comment section.
*****
1. Scally, A. et al. Insights into hominid evolution from the gorilla genome sequence. Nature 483, 169–175 (08 March 2012) doi:10.1038/nature10842

I’m the first author of the gorilla study; thanks for blogging about our research! I thought I might comment on a couple of the points you raise.
You are right that there remains considerable freedom in how we estimate divergence times between the African great ape genera (Homo, Pan and Gorilla) due to our lack of knowledge about ancestral mutation rates. However we do a bit more than simply fit our data to a particular interpretation of the fossil record, because we also point out that recent measurements of the rate in modern humans are low, and we discuss a range of models which could accomodate both this observation and less contentious fossil constraints such as a Miocene divergence for orangutan. Many of the details of this are in the supplement, but one of our goals was to present both genetic and fossil evidence (as in figure 1b) in a manner which couples them more closely and assists further dialogue between paleoanthropology and genetics.
With regard to the particular fossil taxa you mention, some commentators had previously suggested that the genetic evidence precluded their classification as hominins, based on estimates of 4 Mya for the human-chimp speciation. We have shown that for Ardipithecus and Orrorin at least, this is not the case. (There may be other good reasons not to classify them as hominins, but not at present the genetic speciation dates.)
The other point is a technical one about ILS. Actually ILS itself is not necessarily indicative of ‘fuzzy’ or complex speciation, in that we would see it even if there had been clean allopatric splits between the three genera. Instead it’s consistent simply with a large population size in the ancestors of humans and chimps, such that a substantial amount of ancestral variation from the time of gorilla speciation was still present at the time of human-chimp speciation.
This is not to say that the speciation events concerned were not in fact fuzzy. Indeed, given the observations of post-separation genetic exchange in more recent hominid speciations, we speculate that this is not unlikely. It has been suggested before for the human-chimp separation (e.g. Patterson et al., Nature 2006), but as we discuss in the paper, we didn’t actually see unambiguous evidence for this in our analyses.
Thank you for taking the time to stop by, Aylwyn. I have no doubt that with the paper’s release last week you are quite busy.
My statement about the issue of divergence times was meant to highlight the points you raise. As someone who spends more time working with the fossil end of things, it is wonderful when the confidence intervals for genetic divergence times can provide clear evidence to reject certain hypotheses. As you point out, a few years ago it seemed that estimates of the Homo–Pan divergence were moving towards younger dates, possibly providing clearer evidence that some of the earlier putative hominins (Sahelanthropus, Orrorin, possibly Ardipithecus kadaba) did not appear after this divergence. The more recent work attempting to directly estimate mutation rates has obviously started pushing the window back again and thrown a lot of confusion into an already clouded picture (but in a good way, given the independence of empirically derived mutation rates). I like the illustration in figure 1b (which is why I cropped it), and hope that we do see an increase in dialogue between paleoanthropologists and geneticists on this front. Clearly we would be well served by increasing our available fossil sample in the 4-7 Ma time period, in addition to the massive amount of new genetic data which is sure to come down the pipeline in the next few years. I get the sense that the data you present here is unlikely to shift the argument much one way or another on this issue within the paleoanthropological community, as the uncertainty can be argued in a variety of ways to argue for one perspective (i.e. including Sahelanthropus) or another (i.e. excluding Sahelanthropus).
I also apologize for the “fuzziness” of my own use of “fuzziness.” The demographic aspects of speciation, including the effective size of the ancestral population, are part of what I include under my rubric of “fuzziness.” The ancestral effective size is not only a genetic parameter, but also can be construed as an ecological factor associated with the pattern and process of speciation. But the range of effective size estimates available, and the difficulty of uncoupling them from divergence time, allow for multiple speciation models. That we are getting to the point where we can effectively incorporate the fossil and genetic evidence for these kinds of speciation questions, and I think in the later Pliocene and Pleistocene where we have denser fossil and archaeological data sets we are, is amazing. In addition to simply filling in gaps, I see the greatest value in the work of people such as myself who engage in paleoanthropological fieldwork in our ability to produce data that can provide finer calibration on the demographic/ecological underpinnings of genetic models.
Many thanks for posting and good luck!